Researchers from Karolinska Institutet have made a significant breakthrough in understanding how cancer drugs interact with different forms of proteins. Their study, published in Nature Communications, introduces a new method that allows for the distinction of subtle protein variants, known as functional proteoforms. These variations can impact how drugs interact with proteins, influencing the efficacy of treatments and potential side effects.
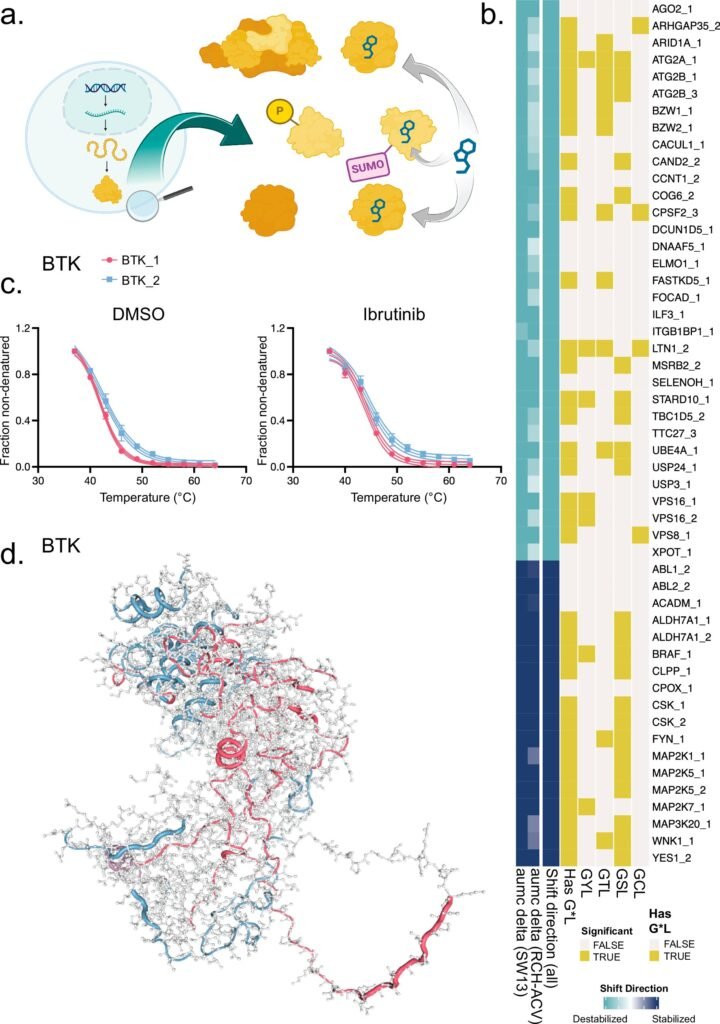
Traditionally, proteins derived from the same gene were considered identical. However, the innovative approach developed by the researchers now enables the identification of unique protein variants that can have varying interactions with drugs. This was demonstrated in the study focusing on ibrutinib, a commonly used drug for certain types of leukemia. The research revealed that ibrutinib interacts differently with distinct variants of the same protein, shedding light on the complexity of drug-proteoform interactions.
The novel method relies on thermal proteomics, where researchers analyze the thermostability of individual protein fragments, or peptides, to distinguish subtle differences between proteoforms. By studying how drugs affect the thermostability of proteins, the researchers were able to identify specific functional proteoforms and their interactions with ibrutinib.
The implications of this research for drug development are profound. Understanding the interactions between functional proteoforms and cancer therapies can significantly impact drug development and precision medicine. The ability to identify these minute details could lead to more effective and personalized treatments in the future.
Moving forward, the research team aims to further refine their method to gain a deeper understanding of how drugs interact with complex protein networks within cells. This advancement is crucial in paving the way for more tailored and efficient treatment strategies in the field of oncology.
For more information on this groundbreaking study, readers can refer to the publication in Nature Communications titled “Functional proteoform group deconvolution reveals a broader spectrum of ibrutinib off-targets” by Isabelle Rose Leo et al. The DOI for the study is 10.1038/s41467-024-54654-8.
This research was conducted at Karolinska Institutet, a leading medical university in Sweden, and underscores the institution’s commitment to advancing scientific knowledge and improving patient care through innovative research initiatives.