Researchers from the Icahn School of Medicine at Mount Sinai in New York, Boston Medical Center, and Boston University Chobanian & Avedisian School of Medicine have collaborated to develop a groundbreaking software platform known as Giotto Suite. This innovative tool aims to simplify the analysis of molecular tissue structures in both healthy and diseased states, as detailed in a recent publication in Nature Methods.
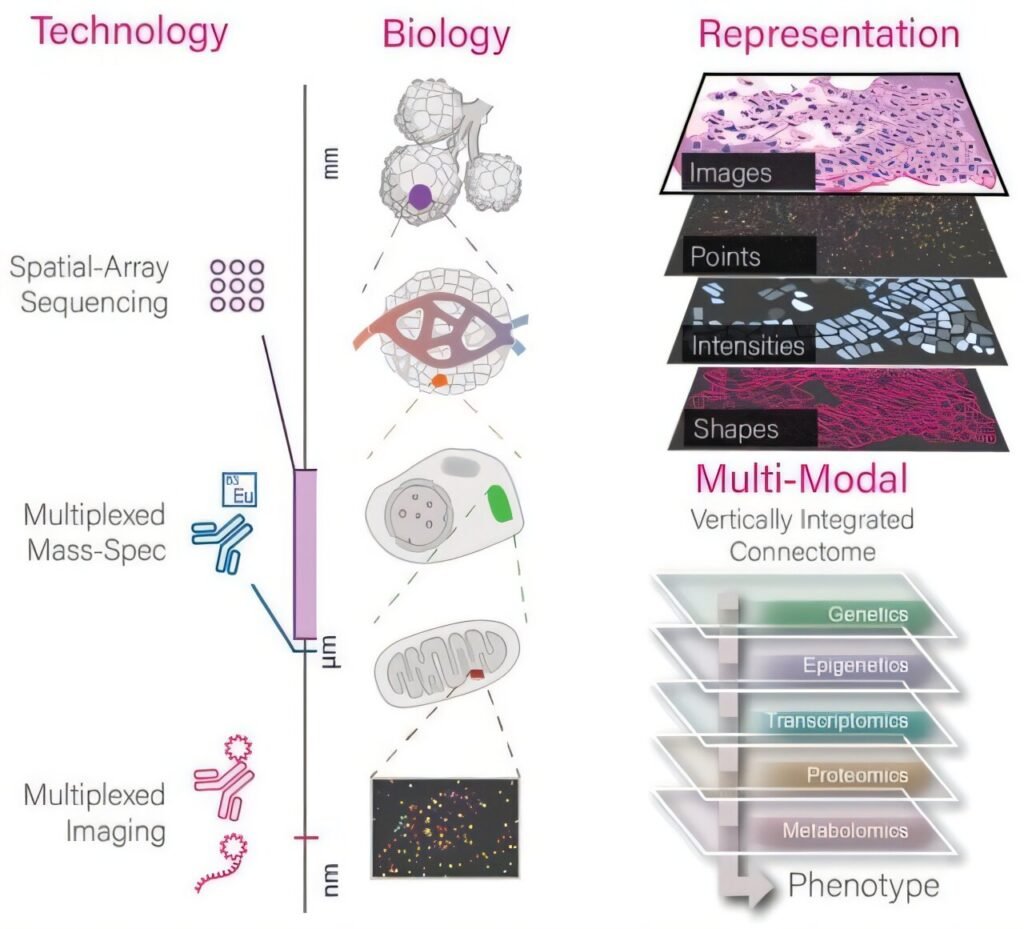
In recent years, advancements in technology have enabled the detailed mapping of RNA and proteins within intact tissues, a field known as spatial omics. These detailed maps provide valuable insights into cellular behavior and interactions within different tissue microenvironments, particularly in the context of diseases such as cancer, neurodegenerative disorders, and immune-related conditions.
According to co-senior corresponding author, Guo-Cheng Yuan, Ph.D., Professor of Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai, the challenge now lies in analyzing and interpreting the vast amount of data generated by spatial omics technologies. The primary focus has shifted from data generation to data analysis, prompting the need for tools that can simplify the complex analytical process and adapt to various types of spatial data and workflows.
To address this need, the research teams developed Giotto Suite, a modular software tool built in the widely-used programming language R. This tool is designed to be compatible with spatial data from any platform, supporting a wide range of analyses including data integration across different measurements and resolutions. Its flexible structure allows users to initiate analysis from multiple points, catering to diverse research needs.
Co-senior corresponding author, Ruben Dries, Ph.D., Assistant Professor of Medicine, Hematology and Medical Oncology at Boston University, highlighted that existing tools often require users to combine multiple packages for a complete analysis. Giotto Suite streamlines this process by consolidating all necessary steps into a single framework, while also ensuring scalability to accommodate the growing complexity of spatial datasets.
The researchers acknowledged that the current version of Giotto Suite may not encompass all available analytical methods, but emphasized their commitment to expanding its features, enhancing interoperability with external software packages, and providing training and support to the research community.
Moving forward, the researchers plan to apply Giotto Suite in their studies of cancer and neurodegenerative diseases, continuously refining the tool based on feedback from the scientific community. The platform is freely accessible to researchers at giottosuite.com, with the hope of fostering broader utilization of spatial omics technologies in biomedical research.
For more information on Giotto Suite, readers can refer to the publication in Nature Methods (DOI: 10.1038/s41592-025-02817-w). This transformative software tool is poised to revolutionize the analysis of complex spatial data across healthy and diseased tissue, empowering researchers to uncover new insights in the field of spatial omics.